Native MRA renal arteries
Native MRA (Magnetic Resonance Angiography) Technique
Native MRA refers to non-contrast MR angiography techniques that utilize intrinsic physiological processes for contrast generation, eliminating the need for gadolinium-based contrast agents. Siemens offers two native MRA techniques under the category of NATIVE: NATIVE TrueFISP and NATIVE SPACE.
NATIVE TrueFISP: NATIVE TrueFISP is based on the TrueFISP (True Fast Imaging with Steady-state Precession) sequence, which is a balanced steady-state gradient echo technique. In this technique, contrast is achieved by using a spatially selective inversion pulse to prepare the imaging volume. This pulse suppresses signal from stationary tissues within the imaging volume and venous blood, enhancing the visibility of arteries. Blood flowing into the imaging volume during the inversion time exhibits high-signal characteristics. This technique can be tailored for arteries or veins by positioning the inversion pulse accordingly. NATIVE TrueFISP can accommodate 3D, 2D, breath-hold, syngo PACE (Prospective Acquisition CorrEction) navigated, and respiratory triggered approaches based on the clinical context.
NATIVE SPACE: NATIVE SPACE is a modified 3D Turbo Spin Echo (TSE) sequence with variable flip angles. It relies on the difference in signal intensity between maximal and minimal blood flow during the cardiac cycle. By subtracting images acquired at different phases of the cardiac cycle, background and venous signals are suppressed, and arterial vasculature becomes more prominent. The sequence enables immediate Inline subtraction and Inline Maximum Intensity Projection (MIP) generation, producing non-invasive MR angiograms in real-time. NATIVE SPACE can also support multi-phase imaging for dynamic angiography in regions like the lower legs.
Clinical Importance and Applications: The development of native MRA techniques like NATIVE TrueFISP and NATIVE SPACE addresses concerns about the use of gadolinium-based contrast agents and offers alternatives for imaging blood vessels. These techniques can be particularly valuable in situations where contrast agent administration is contraindicated or undesirable, such as in patients with renal failure or allergies. By utilizing the inherent flow properties of blood, native MRA provides clinically relevant information about vascular structures without the need for contrast agents, thus enhancing patient safety and reducing healthcare costs.
Challenges and Considerations: While native MRA techniques have advantages, they also have limitations. They may be less effective in cases where physiological processes influencing blood flow are disrupted or altered, such as in patients with severe pathologies. Additionally, patient-specific factors, such as heart rate, respiratory patterns, and overall blood flow, can impact the quality of the images obtained using native MRA techniques. Therefore, careful patient selection and protocol optimization are crucial for achieving high-quality results.
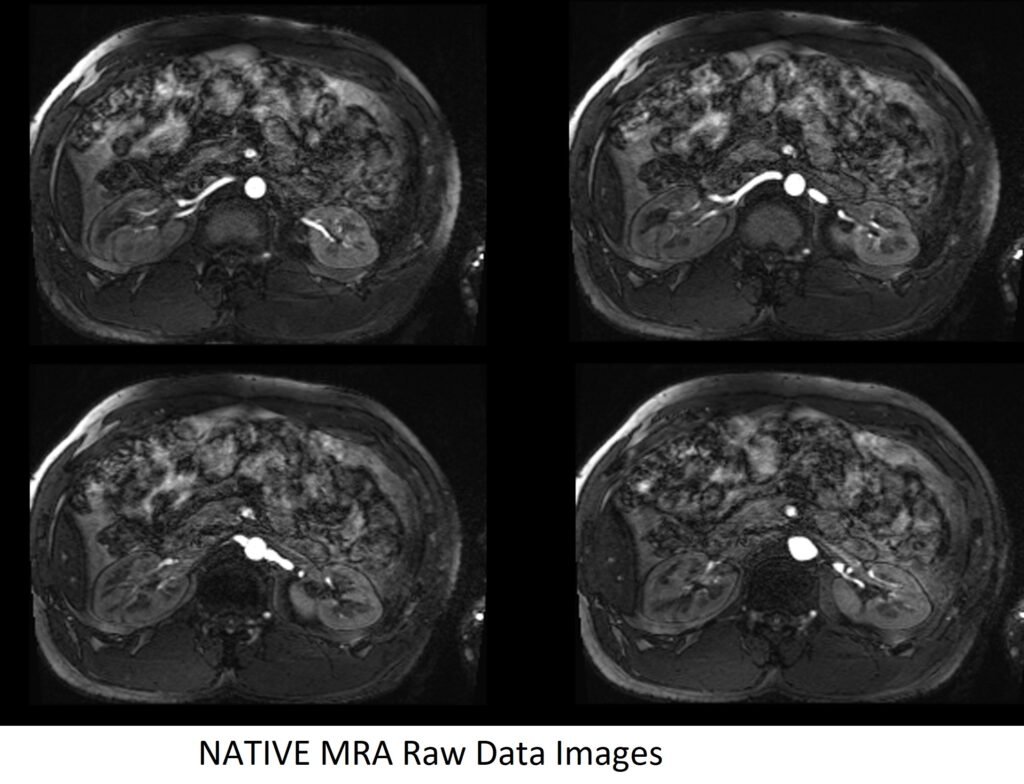
NATIVE MRA Raw Data Images
Indications for magnetic resonance angiography (MRA) renals
- Renal vascular malformation (i.e. arteriovenous malformations and fistulae)
- For the evaluation of renal artery stenosis
- Hypertensive chronic kidney disease
- Atherosclerotic renal artery stenosis
- Fibro muscular dysplasia
- Renal artery aneurysm
- Renal artery dissection
- Renal donors
Contraindications
- Any electrically, magnetically or mechanically activated implant (e.g. cardiac pacemaker, insulin pump biostimulator, neurostimulator, cochlear implant, and hearing aids)
- Intracranial aneurysm clips (unless made of titanium)
- Pregnancy (risk vs benefit ratio to be assessed)
- Ferromagnetic surgical clips or staples
- Metallic foreign body in the eye
- Metal shrapnel or bullet
Patient preparation
- A satisfactory written consent form must be taken from the patient before entering the scanner room
- Ask the patient to remove all metal objects including keys, coins, wallet, cards with magnetic strips, jewellery, hearing aid and hairpins
- Ask the patient to undress and change into a hospital gown
- Instruct the patient to hold their breath for the breath hold scans (its better to coach the patient two to three times before starting the scan)
- If possible provide a chaperone for claustrophobic patients (e.g. relative or staff )
- Offer earplugs or headphones, possibly with music for extra comfort
- Explain the procedure to the patient
- Instruct the patient to keep still
- Note the weight of the patient
Positioning
- Position the patient in supine position with head pointing towards the magnet (head first supine)
- Position the patient's chest over the table respiratory sensor in the spine coil, and place the body coil over the abdomen (from the xiphoid process down to the anterior superior iliac spine).
- Securely tighten the body coil using straps to prevent respiratory artefacts
- Give a pillow under the head and cushions under the legs for extra comfort
- Centre the laser beam localiser over the level of lower intercostal border (i.e. L3 level)
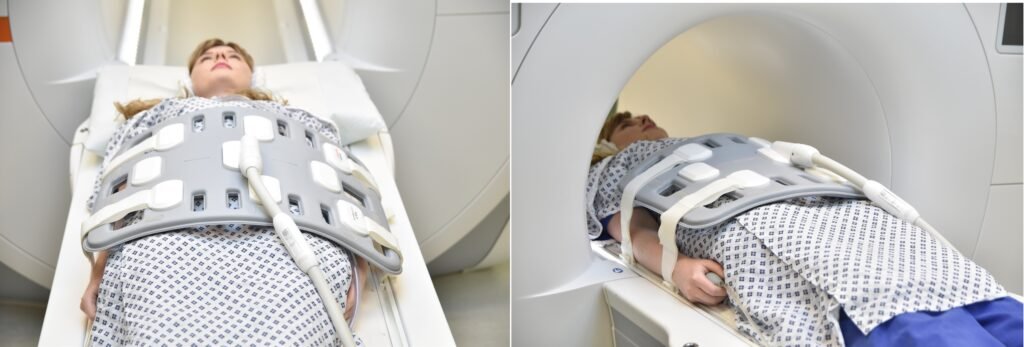
Table sensors
Advanced MRI scanners are equipped with built-in table sensors that detect the respiratory waveform and trigger data acquisition during the expiration phase of the respiratory cycle. Proper patient positioning over the sensor is critical for accurate respiratory gating. This method eliminates the need for external respiratory gating equipment, such as sensors and belts.
If this feature is unavailable on your scanner, please utilize the respiratory bellows. Refer to the manufacturer’s instructions for guidance on using the bellows.

Suggested protocols, parameters and planning
localiser
To localize and plan the sequences, it is essential to acquire a three-plane T2 HASTE localizer initially. These fast single-shot localizers have an acquisition time of under 25 seconds and are highly effective in accurately localizing abdominal structures.

T1 VIBE DIXON 3mm axial BH pre GD(In-opposed phase and water sat)
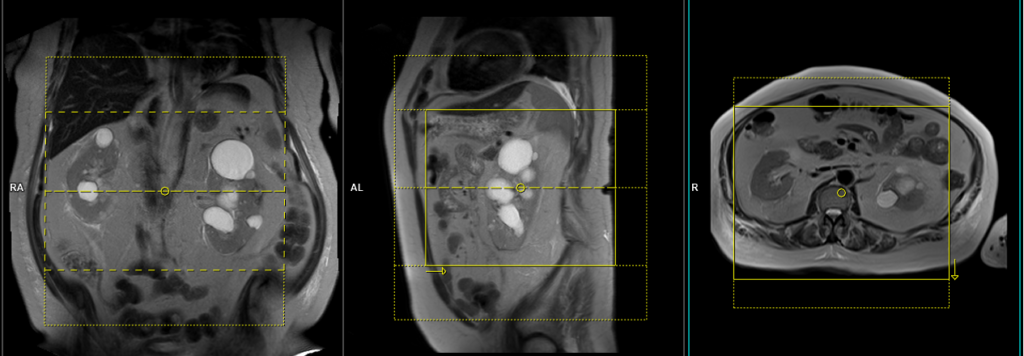
Plan the axial slices on the coronal plane; angle the position block parallel to the right and left renal pelvis. Check the positioning block in the other two planes. An appropriate angle must be given in the sagittal plane (perpendicular to the long axis of kidney). Slices must be sufficient to cover both kidneys from two slices above the upper pole of kidneys down to two slices below the lower pole of kidney. Phase oversampling and, in the case of 3D blocks, slice oversample, must be used to avoid wrap around artefacts. Consider adding saturation bands at the top and bottom of the block to minimize artifacts caused by fat signal, arterial pulsation, and breathing. Instruct the patient to hold their breath during image acquisition.
Parameters
TR 6-7 | TE 2.39 4.77 | FLIP 10 | NXA 1 | SLICE 3 MM | MATRIX 320×320 | FOV 320-350 | PHASE A>P | OVERSAMPLE 20% | BH YES |
T2 tse\haste fat sat breath hold 4mm axial
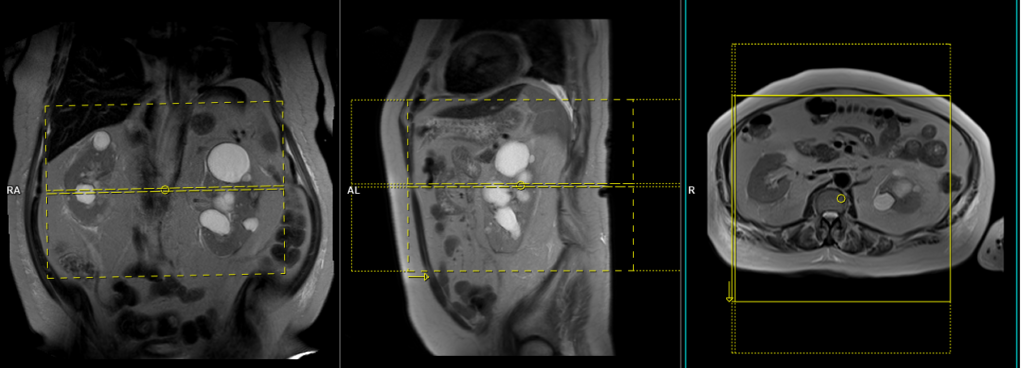
Plan the coronal slices on the axial plane and angle the positioning block parallel to the right and left kidneys. Check the positioning block in the other two planes. Ensure an appropriate angle is given in the sagittal plane (parallel to the long axis of the kidney). The slices should adequately cover both kidneys from the anterior to posterior direction. Phase oversampling should be used to prevent wrap-around artifacts. Consider adding saturation bands at the top and bottom of the block to minimize artifacts caused by fat signal, arterial pulsation, and breathing. Instruct the patient to hold their breath during image acquisition.
Parameters HASTE FAT SAT
TR 2000-2500 | TE 90-110 | FAT SAT SPAIR | NEX 1 | SLICE 4MM | MATRIX 320×256 | FOV 300 | PHASE R>L | OVERSAMPLE 50% | TRIGGER NO |
Parameters T2 FAT SAT
| TR6000-7000 | TE 150 | FAT SAT SPAIR | NEX 1 | SLICE 4MM | MATRIX 256×208 | FOV 280-300 | PHASE A>P | OVERSAMPLE 80% | IPAT ON |
NATIVE TrueFISP axial respiratory gated 1mm
This particular sequence is performed using a table sensor respiratory-gated sequence. Please choose the appropriate sequence from the protocol.
Plan the axial native block on the coronal plane by first carefully positioning the imaging Field of View (FOV). Ensure that the upper border of the FOV is placed very close to the origin of the targeted vessel. Avoid incorporating an excessive amount of the proximal aorta within the FOV. Now, adjust the positioning block’s angle to run parallel with both the right and left renal pelvis.
To ensure accurate positioning, verify the positioning block in the other two planes as well. It’s essential to maintain an appropriate angle in the sagittal plane, ensuring alignment horizontally across the kidney. For an enhanced inflow effect, make sure that the upper limit of the inversion band aligns precisely with the upper FOV limit. This alignment prevents any loss of signal originating from the inflowing arterial blood.
For improved accuracy and to avoid venous contamination, it’s highly recommended to employ two inversion bands. The upper band should have an inversion time (TI) of 1300-1400ms, while the lower band should possess a TI of 800ms.
The acquisition time for the scan typically falls within a range of 2 to 5 minutes. However, this duration may vary based on factors such as the patient’s breathing cycle. Throughout the scan, diligently monitor the respiratory sensor signal. This ongoing observation ensures the successful completion of the scan and helps uphold image quality standards.
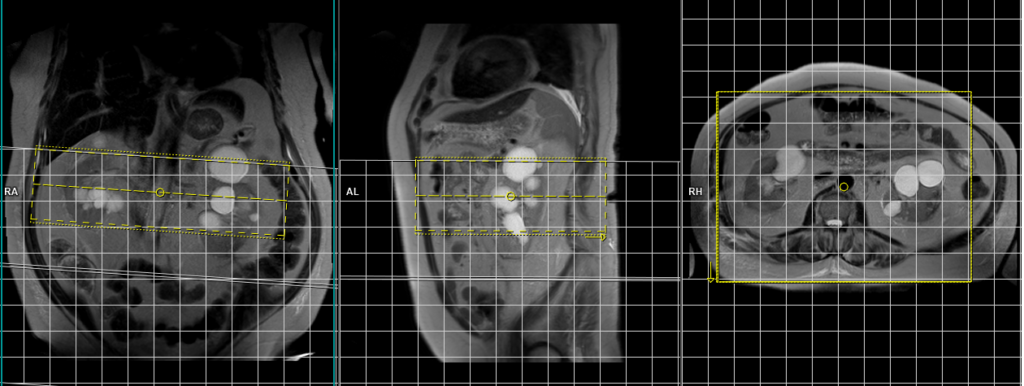
Parameters
TR 2000-3000 | TE 1.5 2 | FLIP 90 | NXA 1 | SLICE 1 MM | MATRIX 256×256 | FOV 320-350 | PHASE A>P | OVERSAMPLE 10% | IPAT YES |
MIP (Maximum Intensity Projection) of NATIVE MRA raw data
MIP (Maximum Intensity Projection) reconstruction is a technique used in MRI (Magnetic Resonance Imaging) to create a two-dimensional image that displays the maximum intensity along a chosen projection path. It involves projecting the highest signal intensity voxel from each slice along a specific viewing direction onto a single image plane. MIP reconstructions are commonly used in vascular imaging to enhance the visualization of blood vessels and highlight areas of high signal intensity. This technique allows for a comprehensive overview of the vasculature and aids in identifying abnormalities or areas of interest in a three-dimensional volume dataset.

